
Genetic Engineering & Biotechnology News (GEN) is the world's most widely read biotech publication. It provides the R&D; community with critical information on the tools, technologies, and trends that drive the biotech industry.
Issue link: http://gen.epubxp.com/i/145822
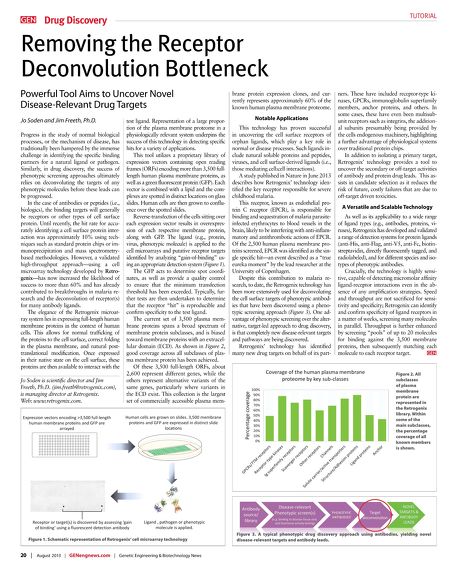
TUTORIAL Drug Discovery Removing the Receptor Deconvolution Bottleneck Powerful Tool Aims to Uncover Novel Disease-Relevant Drug Targets Jo Soden and Jim Freeth, Ph.D. Progress in the study of normal biological processes, or the mechanism of disease, has traditionally been hampered by the immense challenge in identifying the specifc binding partners for a natural ligand or pathogen. Similarly, in drug discovery, the success of phenotypic screening approaches ultimately relies on deconvoluting the targets of any phenotypic molecules before these leads can be progressed. In the case of antibodies or peptides (i.e., biologics), the binding targets will generally be receptors or other types of cell surface protein. Until recently, the hit rate for accurately identifying a cell surface protein interaction was approximately 10% using techniques such as standard protein chips or immunoprecipitation and mass spectrometrybased methodologies. However, a validated high-throughput approach—using a cell microarray technology developed by Retrogenix—has now increased the likelihood of success to more than 60% and has already contributed to breakthroughs in malaria research and the deconvolution of receptor(s) for many antibody ligands. The elegance of the Retrogenix microarray system lies in expressing full-length human membrane proteins in the context of human cells. This allows for normal traffcking of the proteins to the cell surface, correct folding in the plasma membrane, and natural posttranslational modifcation. Once expressed in their native state on the cell surface, these proteins are then available to interact with the Jo Soden is scientifc director and Jim Freeth, Ph.D. (jim.freeth@retrogenix.com), is managing director at Retrogenix. Web: www.retrogenix.com. test ligand. Representation of a large proportion of the plasma membrane proteome in a physiologically relevant system underpins the success of this technology in detecting specifc hits for a variety of applications. This tool utilizes a proprietary library of expression vectors containing open reading frames (ORFs) encoding more than 3,500 fulllength human plasma membrane proteins, as well as a green fuorescent protein (GFP). Each vector is combined with a lipid and the complexes are spotted in distinct locations on glass slides. Human cells are then grown to confuence over the spotted slides. Reverse-transfection of the cells sitting over each expression vector results in overexpression of each respective membrane protein, along with GFP. The ligand (e.g., protein, virus, phenotypic molecule) is applied to the cell microarrays and putative receptor targets identifed by analyzing "gain-of-binding" using an appropriate detection system (Figure 1). The GFP acts to determine spot coordinates, as well as provide a quality control to ensure that the minimum transfection threshold has been exceeded. Typically, further tests are then undertaken to determine that the receptor "hit" is reproducible and confrm specifcity to the test ligand. The current set of 3,500 plasma membrane proteins spans a broad spectrum of membrane protein subclasses, and is biased toward membrane proteins with an extracellular domain (ECD). As shown in Figure 2, good coverage across all subclasses of plasma membrane protein has been achieved. Of these 3,500 full-length ORFs, about 2,600 represent different genes, while the others represent alternative variants of the same genes, particularly where variants in the ECD exist. This collection is the largest set of commercially accessible plasma mem- Figure 1. Schematic representation of Retrogenix' cell microarray technology 20 | August 2013 | GENengnews.com | Genetic Engineering & Biotechnology News brane protein expression clones, and currently represents approximately 60% of the known human plasma membrane proteome. Notable Applications This technology has proven successful in uncovering the cell surface receptors of orphan ligands, which play a key role in normal or disease processes. Such ligands include natural soluble proteins and peptides, viruses, and cell surface-derived ligands (i.e., those mediating cell:cell interactions). A study published in Nature in June 2013 describes how Retrogenix' technology identifed the key receptor responsible for severe childhood malaria. This receptor, known as endothelial protein C receptor (EPCR), is responsible for binding and sequestration of malaria parasiteinfected erythrocytes to blood vessels in the brain, likely to be interfering with anti-infammatory and antithrombotic actions of EPCR. Of the 2,500 human plasma membrane proteins screened, EPCR was identifed as the single specifc hit—an event described as a "true eureka moment" by the lead researcher at the University of Copenhagen. Despite this contribution to malaria research, to date, the Retrogenix technology has been more extensively used for deconvoluting the cell surface targets of phenotypic antibodies that have been discovered using a phenotypic screening approach (Figure 3). One advantage of phenotypic screening over the alternative, target-led approach to drug discovery, is that completely new disease-relevant targets and pathways are being discovered. Retrogenix' technology has identifed many new drug targets on behalf of its part- ners. These have included receptor-type kinases, GPCRs, immunoglobulin superfamily members, anchor proteins, and others. In some cases, these have even been multisubunit receptors such as integrins, the additional subunits presumably being provided by the cells endogenous machinery, highlighting a further advantage of physiological systems over traditional protein chips. In addition to isolating a primary target, Retrogenix' technology provides a tool to uncover the secondary or off-target activities of antibody and protein drug leads. This assists in candidate selection as it reduces the risk of future, costly failures that are due to off-target driven toxicities. A Versatile and Scalable Technology As well as its applicability to a wide range of ligand types (e.g., antibodies, proteins, viruses), Retrogenix has developed and validated a range of detection systems for protein ligands (anti-His, anti-Flag, anti-V5, anti-Fc, biotinstreptavidin, directly fuorescently tagged, and radiolabeled), and for different species and isotypes of phenotypic antibodies. Crucially, the technology is highly sensitive, capable of detecting micromolar affnity ligand-receptor interactions even in the absence of any amplifcation strategies. Speed and throughput are not sacrifced for sensitivity and specifcity; Retrogenix can identify and confrm specifcity of ligand receptors in a matter of weeks, screening many molecules in parallel. Throughput is further enhanced by screening "pools" of up to 20 molecules for binding against the 3,500 membrane proteins, then subsequently matching each molecule to each receptor target. Figure 2. All subclasses of plasma membrane protein are represented in the Retrogenix library. Within some of the main subclasses, the percentage coverage of all known members is shown. Figure 3. A typical phenotypic drug discovery approach using antibodies, yielding novel disease-relevant targets and antibody leads.